Our lab conducts research in image analysis, statistical shape modeling, and machine learning to discover the underlying mechanisms of diseases. In particular, we develop novel methods for image registration, image segmentation, and population-based geometric shape analysis. Our research has potential applications in noninvasive disease diagnosis, screening, and treatment.
Publications
Check Google Scholar Citation for full publications.
Ph.D. Dissertation
Miaomiao Zhang, Bayesian Models On Manifolds For Image Registration and Statistical Shape Analysis
-> Defense Video
Jian Wang, Deep Image Analysis Based on Deformable Shapes and Its Applications to Neuroimaging
-> Defense Video
Jiarui (Jerry) Xing, Deep Motion Networks to Advance Cardiac Magnetic Resonance Strain Analysis
-> Defense Video
Projects
Fast image registration FLASH (Fourier-approximated Lie Algebras for SHooting) [Paper I ] [Paper II ] [Code ]
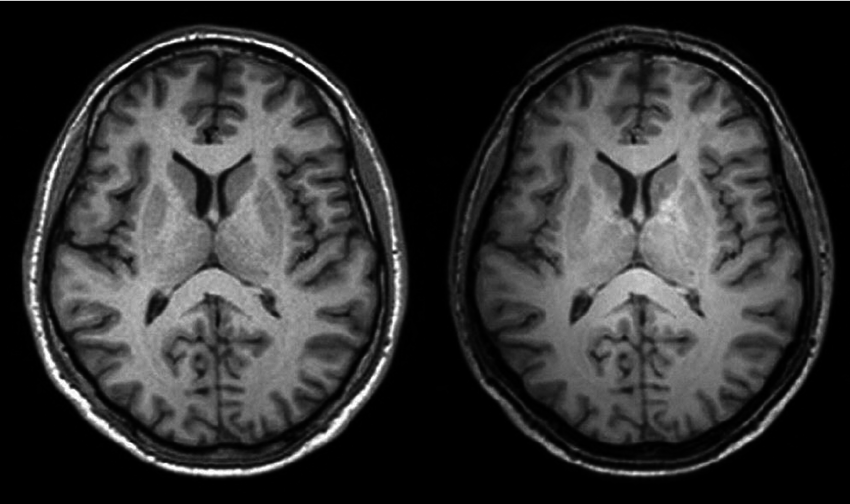 |
This is a ultrafast implementation of LDDMM (large deformation diffeomorphic metric mapping) with geodesic shooting algorithm for deformable image registration. Our algorithm dramatically speeds up the state-of-art registration methods with little to no loss of accuracy. |
Registration Uncertainty Quantification [Paper ]
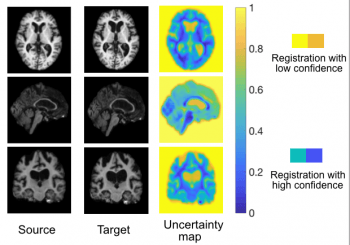 |
This project develops efficient algorithms to quantify the uncertainty of registration results. This is critical to fair assessment on the final estimated transformations and subsequent improvement on the accuracy of predictive registration models. Our algorithm improves the reliability of registration in clinical applications, e.g., real-time image guided navigation system for neurosurgery. |
Motion correction for placental DW-MRI scans
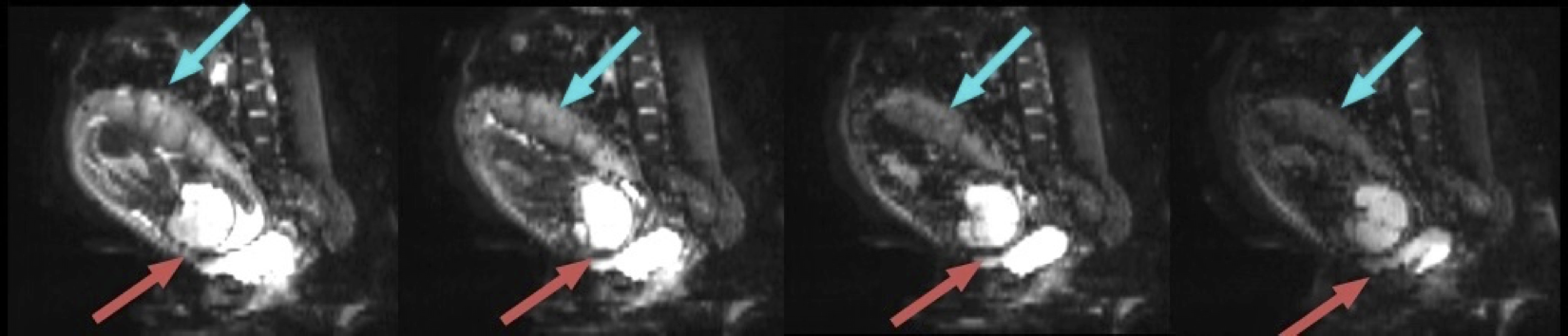 |
Placental pathology, such as immune cell infiltration and inflammation, is a common reason for preterm labor. It occurs in around 11 percent of world pregnancies. The goal of this project is to develop robust computational models to monitor placental health through in-Utero diffusion-weighted MR images (DW-MRI). More specifically, we are keen to design methods that correct large motion artifacts caused by maternal breathing and fetal movements for severely noise-corrupted 3D placental images. |
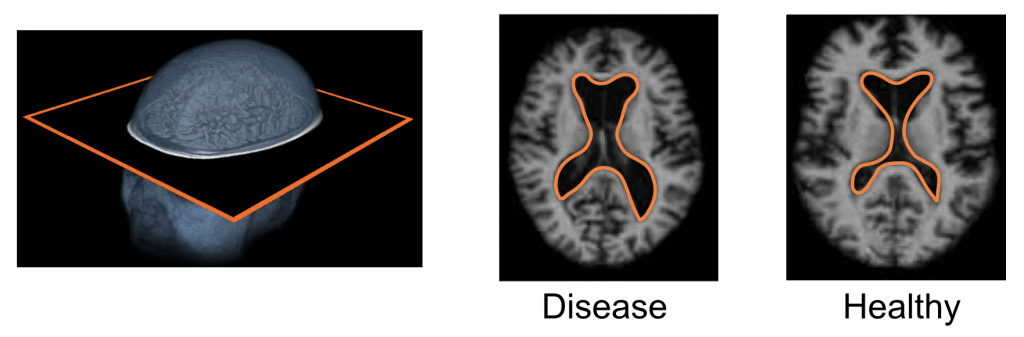
Population-based Anatomical Shape Analysis
 |

