MIA Lab

Our lab conducts research in image analysis, statistical shape modeling, and machine learning to discover the underlying
mechanisms of diseases. In particular, we develop novel methods for image registration, image segmentation, and
population-based geometric shape analysis. Our research has potential applications in noninvasive disease diagnosis,
screening, and treatment.
Publications
Ph.D. Dissertation
Projects
MGAug: Multimodal Geometric Augmentation in Latent Spaces of Image Deformations
[Paper ]
[Code ]

Geometric transformations have been widely used to augment the size of training images.
Existing methods often assume a unimodal distribution of the underlying transformations between images, which limits their
power when data with multimodal distributions occur. In this paper, we propose a novel model, Multimodal Geometric Augmentation (MGAug),
that for the first time generates augmenting transformations in a multimodal latent space of geometric deformations.
Invariant Shape Representation Learning For Image Classification
[Paper ]
[Code ]

Geometric shape features have been widely used as strong predictors for image classification. Nevertheless, most existing classifiers
such as deep neural networks (DNNs) directly leverage the statistical correlations between these shape features and target variables.
However, these correlations can often be spurious and unstable across different environments (e.g., in different age groups, certain types of
brain changes have unstable relations with neurodegenerative disease); hence le,mading to biased or inaccurate predictions. In this paper,
we introduce a novel framework that for the first time develops invariant shape representation learning (ISRL) to further strengthen the robustness
of image classifiers. In contrast to existing approaches that mainly derive features in the image space, our model ISRL is designed to jointly
capture invariant features in latent shape spaces parameterized by deformable transformations.
ToRL: Topology-preserving Representation Learning Of Object Deformations From Images
[Paper ]
[Code ]

Representation learning of object deformations from images has been a long-standing challenge in various image or video analysis tasks.
Existing deep neural networks typically focus on visual features (e.g., intensity and texture), but they often fail to capture the underlying
geometric and topological structures of objects. This limitation becomes especially critical in areas, such as medical imaging and 3D modeling,
where maintaining the structural integrity of objects is essential for accuracy and generalization across diverse datasets. In this paper,
we introduce ToRL, a novel {\em Topology-preserving Representation Learning} model that, for the first time, offers an explicit mechanism for
modeling intricate object topology in the latent feature space. We develop a comprehensive learning framework that captures object deformations
via learned transformation groups in the latent space. Each layer of our network's decoder is carefully designed with an integrated smooth composition
module, ensuring that topological properties are preserved throughout the learning process. Moreover, in contrast to a few related works that rely
on a reference image to predict object deformations during inference, our approach eliminates this impractical requirement.
Multimodal Deep Learning to Differentiate Tumor Recurrence from Treatment Effect in Human Glioblastoma
[Paper ]

Differentiating tumor progression (TP) or recurrence from treatment-related necrosis (TN) is critical for clinical management decisions in glioblastoma (GBM).
Dynamic FDG PET (dPET), an advance from traditional static FDG PET, may prove advantageous in clinical staging. dPET includes novel methods of a model-corrected
blood input function that accounts for partial volume averaging to compute parametric maps that reveal kinetic information. In a preliminary study, a convolution
neural network (CNN) was trained to predict classification accuracy between TP and TN for $35$ brain tumors from $26$ subjects in the PET-MR image space.
3D parametric PET Ki (from dPET), traditional static PET standardized uptake values (SUV), and also the brain tumor MR voxels formed the input for the CNN.
The average test accuracy across all leave-one-out cross-validation iterations adjusting for class weights was $0.56$ using only the MR, $0.65$ using only the SUV,
and $0.71$ using only the Ki voxels. Combining SUV and MR voxels increased the test accuracy to $0.62$. On the other hand, MR and Ki voxels increased the test
accuracy to $0.74$. Thus, dPET features alone or with MR features in deep learning models would enhance prediction accuracy in differentiating TP vs TN in GBM.
SADIR: Shape-Aware Diffusion Models for 3D Image Reconstruction
[Paper ]
[Code ]

3D image reconstruction from a limited number of 2D images has been a long-standing challenge in computer vision and image analysis. While deep learning-based
approaches have achieved impressive performance in this area, existing deep networks often fail to effectively utilize the shape structures of objects presented
in images. As a result, the topology of reconstructed objects may not be well preserved, leading to the presence of artifacts such as discontinuities, holes,
or mismatched connections between different parts. In this paper, we propose a shape-aware network based on diffusion models for 3D image reconstruction,
named SADIR, to address these issues. In contrast to previous methods that primarily rely on spatial correlations of image intensities for 3D reconstruction,
our model leverages shape priors learned from the training data to guide the reconstruction process.
Diffusion Models To Predict 3D Late Mechanical Activation From Sparse 2D Cardiac MRIs
[Paper ]
[Code ]
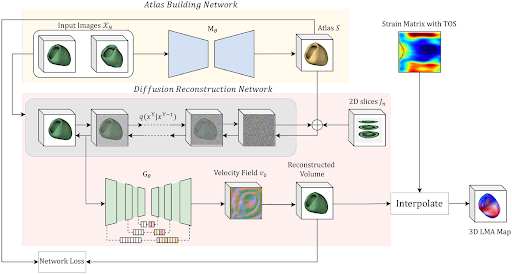
Identifying regions of late mechanical activation (LMA) of the left ventricular (LV) myocardium is critical in determining the optimal pacing site for cardiac
resynchronization therapy in patients with heart failure. Several deep learning-based approaches have been developed to predict 3D LMA maps of LV myocardium from
a stack of sparse 2D cardiac magnetic resonance imaging (MRIs). However, these models often loosely consider the geometric shape structure of the myocardium.
This makes the reconstructed activation maps suboptimal; hence leading to a reduced accuracy of predicting the late activating regions of hearts. In this paper,
we propose to use shape-constrained diffusion models to better reconstruct a 3D LMA map, given a limited number of 2D cardiac MRI slices. In contrast to previous
methods that primarily rely on spatial correlations of image intensities for 3D reconstruction, our model leverages object shape as priors learned from the training
data to guide the reconstruction process.
Deep learning to predict myocardial scar burden and uncertainty quantification
[Paper ]

Late gadolinium enhancement (LGE) is a defining feature of cardiac magnetic resonance imaging (CMR) that provides critical diagnostic and prognostic value for
patients with underlying cardiomyopathy. However, interpretation of LGE can be challenging with significant interobserver variability. In this methodology study,
we applied a deep learning model to automatically identify LGE in patients with known scar and to quantify the degree of uncertainty.
TLRN: Temporal Latent Residual Networks for Large Deformation Image Registration
[Paper ]
[Code ]

This paper presents a novel approach, termed {\em Temporal Latent Residual Network (TLRN)}, to predict a sequence of deformation fields in time-series image registration.
The challenge of registering time-series images often lies in the occurrence of large motions, especially when images differ significantly from a reference
(e.g., the start of a cardiac cycle compared to the peak stretching phase).
Our code is publicly available at
https://github.com/nellie689/TLRN .
IGG: Image Generation Informed by Geodesic Dynamics in Deformation Spaces

Generative models have recently gained increasing attention in image generation and editing tasks. However, they often lack a direct connection to object geometry,
which is crucial in sensitive domains such as computational anatomy, biology, and robotics. This paper presents a novel framework for Image Generation informed by
Geodesic dynamics (IGG) in deformation spaces. Our IGG model comprises two key components: (i) an efficient autoencoder that explicitly learns the geodesic path of
image transformations in the latent space; and (ii) a latent geodesic diffusion model that captures the distribution of latent representations of geodesic deformations
conditioned on text instructions. By leveraging geodesic paths, the method ensures smooth, topology-preserving, and interpretable deformations, capturing complex
variations in image structures while maintaining geometric consistency.
Our code will be publicly available at Github.
Neurepdiff: Neural operators to predict geodesics in deformation spaces
[Paper ]
[Code ]
.png)
This paper presents NeurEPDiff, a novel network to fast predict the geodesics in deformation spaces generated by a well known Euler-Poincaré differential equation (EPDiff).
LaMoD: Latent Motion Diffusion Model For Myocardial Strain Generation
[Paper ]
[Code ]
Motion and deformation analysis of cardiac magnetic resonance (CMR) imaging videos is crucial for assessing myocardial strain of patients with abnormal heart functions.
Recent advances in deep learning-based image registration algorithms have shown promising results in predicting motion fields from routinely acquired CMR sequences.
However, their accuracy often diminishes in regions with subtle appearance changes, with errors propagating over time. Advanced imaging techniques, such as displacement
encoding with stimulated echoes (DENSE) CMR, offer highly accurate and reproducible motion data but require additional image acquisition, which poses challenges in busy
clinical flows. In this paper, we introduce a novel Latent Motion Diffusion model (LaMoD) to predict highly accurate DENSE motions from standard CMR videos. More specifically,
our method first employs an encoder from a pre-trained registration network that learns latent motion features (also considered as deformation-based shape features) from
image sequences. Supervised by the ground-truth motion provided by DENSE, LaMoD then leverages a probabilistic latent diffusion model to reconstruct accurate motion from
these extracted features.
https://github.com/jr-xing/LaMoD .
Scar-aware Late Mechanical Activation Detection Network For Optimal Cardiac resynchronization therapy Planning
[Paper ]

Accurate identification of late mechanical activation (LMA) regions is crucial for optimal cardiac resynchronization therapy (CRT) lead implantation.
However, existing approaches using cardiac magnetic resonance (CMR) imaging often overlook myocardial scar information, which may be mistakenly identified
as delayed activation regions. To address this issue, we propose a scar-aware LMA detection network that simultaneously detects myocardial scar and prevents
LMA localization in these scarred regions. More specifically, our model integrates a pre-trained scar segmentation network using late gadolinium enhancement
(LGE) CMRs into a LMA detection network based on highly accurate strain derived from displacement encoding with stimulated echoes (DENSE) CMRs.
Multimodal Learning to Improve Cardiac Late Mechanical Activation Detection From Cine MR Images
[Paper ]
[Code ]

This paper presents a multimodal deep learning framework that utilizes advanced image techniques to improve the performance of clinical analysis heavily dependent
on routinely acquired standard images. More specifically, we develop a joint learning network that for the first time leverages the accuracy and reproducibility of
myocardial strains obtained from Displacement Encoding with Stimulated Echo (DENSE) to guide the analysis of cine cardiac magnetic resonance (CMR) imaging in late
mechanical activation (LMA) detection. An image registration network is utilized to acquire the knowledge of cardiac motions, an important feature estimator of
strain values, from standard cine CMRs.
Joint Deep Learning for Improved Myocardial Scar Detection
[Paper ]
[Code ]

Automated identification of myocardial scar from late gadolinium enhancement cardiac magnetic resonance images (LGE-CMR) is limited by image noise and artifacts
such as those related to motion and partial volume effect. This paper presents a novel joint deep learning (JDL) framework that improves such tasks by utilizing
simultaneously learned myocardium segmentations to eliminate negative effects from non-region-of-interest areas. Our method introduces a message passing module
where myocardium segmentation information directly guides scar detectors, leading to improved performance compared to state-of-the-art methods.
Multitask Learning for Improved Late Mechanical Activation Detection of Heart from Cine Dense MRI
[Paper ]
[Code ]
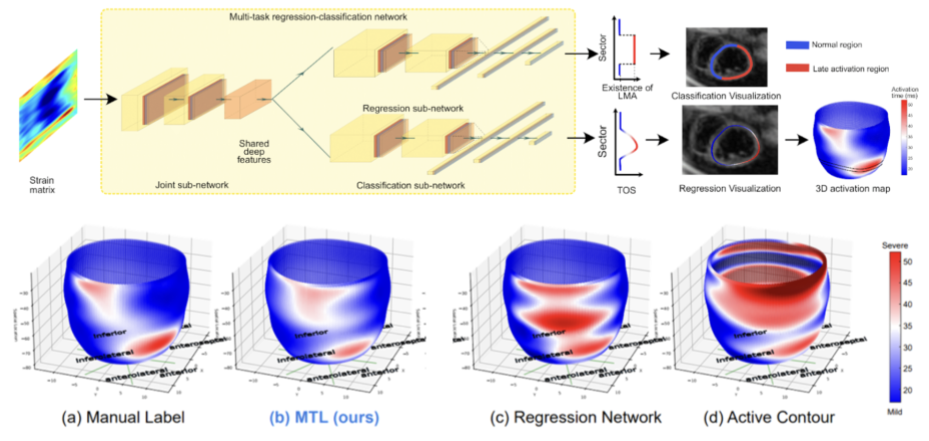
The selection of an optimal pacing site, which is ideally scar-free and late activated, is critical to the response of cardiac resynchronization therapy (CRT).
Despite the success of current approaches formulating the detection of such late mechanical activation (LMA) regions as a problem of activation time regression,
their accuracy remains unsatisfactory, particularly in cases where myocardial scar exists. To address this issue, this paper introduces a multi-task deep learning
framework that simultaneously estimates LMA amount and classifies the scar-free LMA regions based on cine displacement encoding with stimulated echoes (DENSE)
magnetic resonance imaging (MRI). With a newly introduced auxiliary LMA region classification sub-network, our proposed model shows more robustness to the complex
pattern caused by myocardial scar, significantly eliminates their negative effects in LMA detection, and in turn improves the performance of scar classification.
Deep Networks to Automatically Detect Late-Activating Regions of the Heart
[Paper]
[Code]

This paper presents a novel method to automatically identify late-activating regions of the left ventricle from cine Displacement Encoding with Stimulated Echo (DENSE)
MR images. We develop a deep learning framework that identifies late mechanical activation in heart failure patients by detecting the Time to the Onset of circumferential
Shortening (TOS). In particular, we build a cascade network performing end-to-end (i) segmentation of the left ventricle to analyze cardiac function, (ii) prediction of
TOS based on spatiotemporal circumferential strains computed from displacement maps, and (iii) 3D visualization of delayed activation maps. Our approach results in dramatic
savings of manual labors and computational time over traditional optimization-based algorithms.
TPIE: Topology-Preserved Image Editing With Text Instructions
Preserving topological structures is important in real-world applications, particularly in sensitive domains such as healthcare and medicine, where the correctness of human
anatomy is critical. However, most existing image editing models focus on manipulating intensity and texture features, often overlooking object geometry within images.
To address this issue, this paper introduces a novel method, Topology-Preserved Image Editing with text instructions (TPIE), that for the first time ensures the topology and
geometry remaining intact in edited images through text-guided generative diffusion models. More specifically, our method treats newly generated samples as deformable variations
of a given input template, allowing for controllable and structure-preserving edits.
Fast image registration FLASH (Fourier-approximated Lie Algebras for SHooting)
[Paper I ]
[Paper II ]
[Code ]

This is a ultrafast implementation of LDDMM (large deformation diffeomorphic metric mapping) with geodesic shooting algorithm for deformable image registration.
Our algorithm dramatically speeds up the state-of-art registration methods with little to no loss of accuracy.
Registration Uncertainty Quantification
[Paper ]

This project develops efficient algorithms to quantify the uncertainty of registration results.
This is critical to fair assessment on the final estimated transformations and subsequent improvement on the accuracy of predictive registration models.
Our algorithm improves the reliability of registration in clinical applications, e.g., real-time image guided navigation system for neurosurgery.
Motion correction for placental DW-MRI scans

Placental pathology, such as immune cell infiltration and inflammation, is a common reason for preterm labor. It occurs in around 11 percent of world pregnancies.
The goal of this project is to develop robust computational models to monitor placental health through in-Utero diffusion-weighted MR images (DW-MRI).
More specifically, we are keen to design methods that correct large motion artifacts caused by maternal breathing and fetal movements for severely noise-corrupted 3D placental images.
Population-based Anatomical Shape Analysis
3D brain tumor segmentation









.png)














