| Category |
Representative Papers |
Overall |
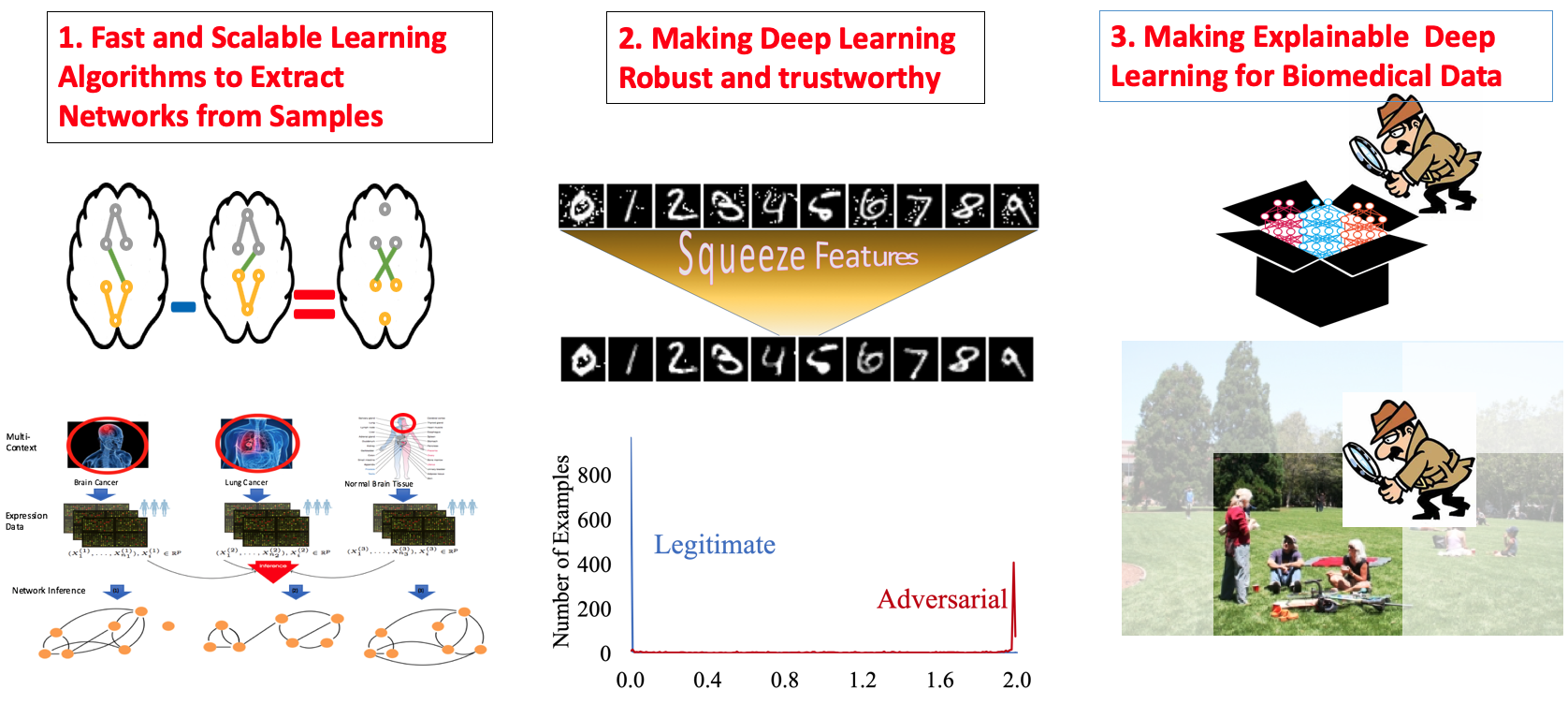
|
Trustworthy Machine Learning
[ Our tools for making machine learning robust and trustworthy]
|
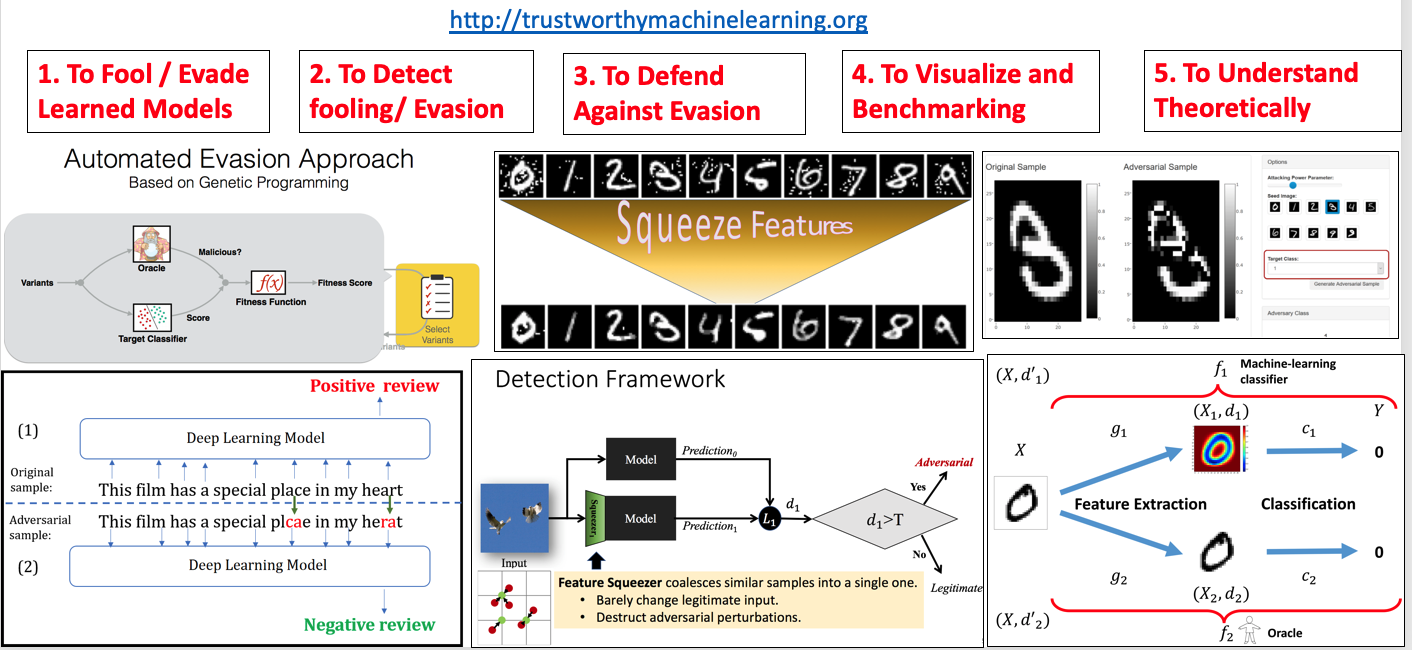
- "Feature Squeezing: Detecting Adversarial Examples in Deep Neural Networks",NDSS 2018 (PDF)
- "Black-box Generation of Adversarial Text Sequences to Evade Deep Learning Classifiers", 2018 IEEE Security and Privacy Workshops (SPW), Ji Gao, Jack Lanchantin, Mary Lou Soffa, Yanjun Qi, (PDF)
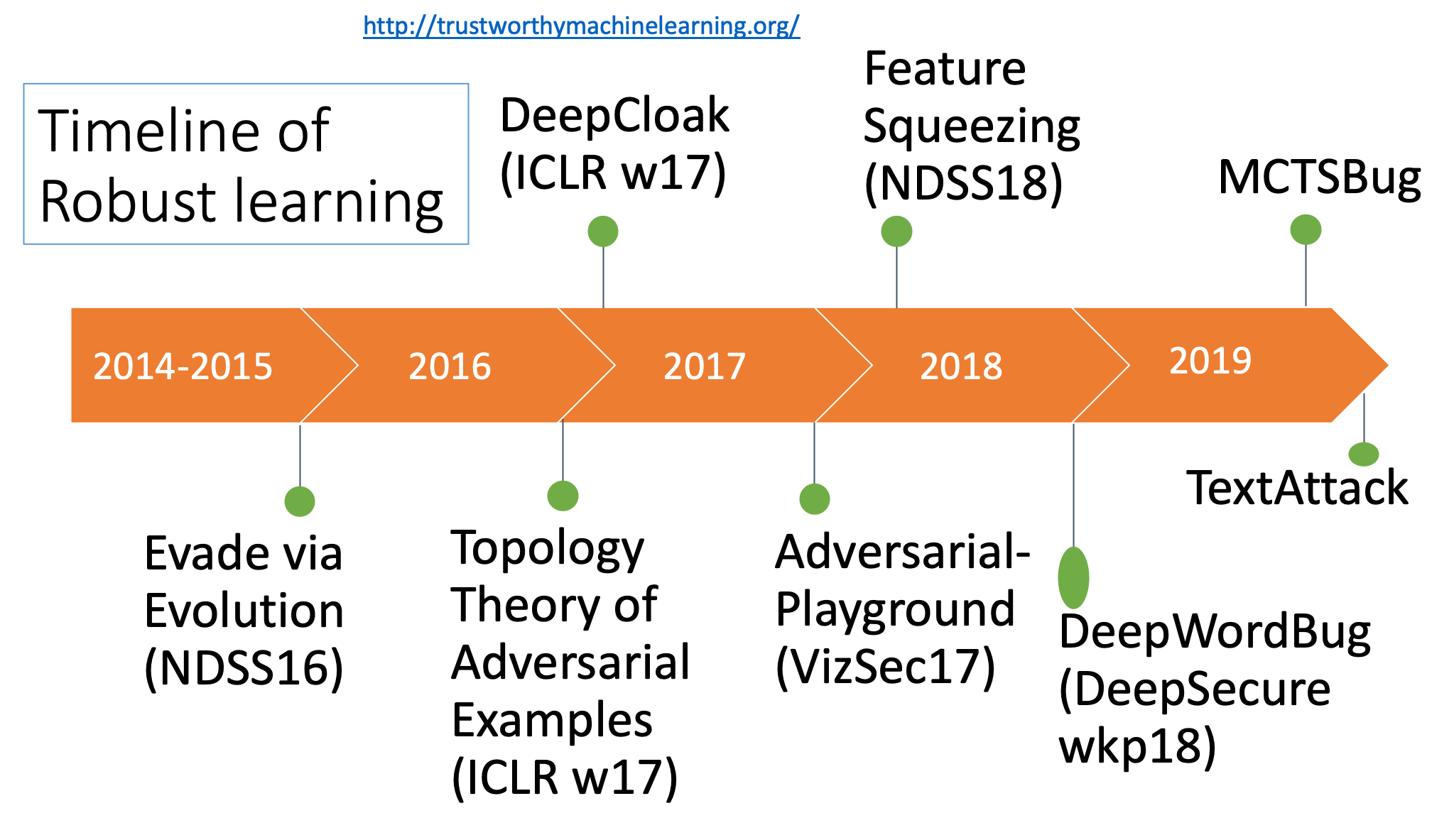
- "Automatically Evading Classifiers",NDSS 2016 (PDF)(Talk)
- "A Theoretical Framework for Robustness of (Deep) Classifiers Against Adversarial Samples", ICLR-17 (Arxiv)
- "Feature Squeezing: Detecting Adversarial Examples in Deep Neural Networks", (Arxiv)
- "DeepCloak: Masking Deep Neural Network Models for Robustness against Adversarial Samples",ICLR-17 workshop (Arxiv)
- "Adversarial Playground: A Visualization Suite for Adversarial Sample Generation", Norton, Andrew and Qi, Yanjun,
(Arxiv)
|
(Deep) Representation Learning on sequential data
(genome or epigenomic data, product review text, bio-reports text, protein sequence strings, etc.)
[ Our Tools for Mining Sequential Bio-Data]
[ Our deep Learning tools on Discrete Structures]
|
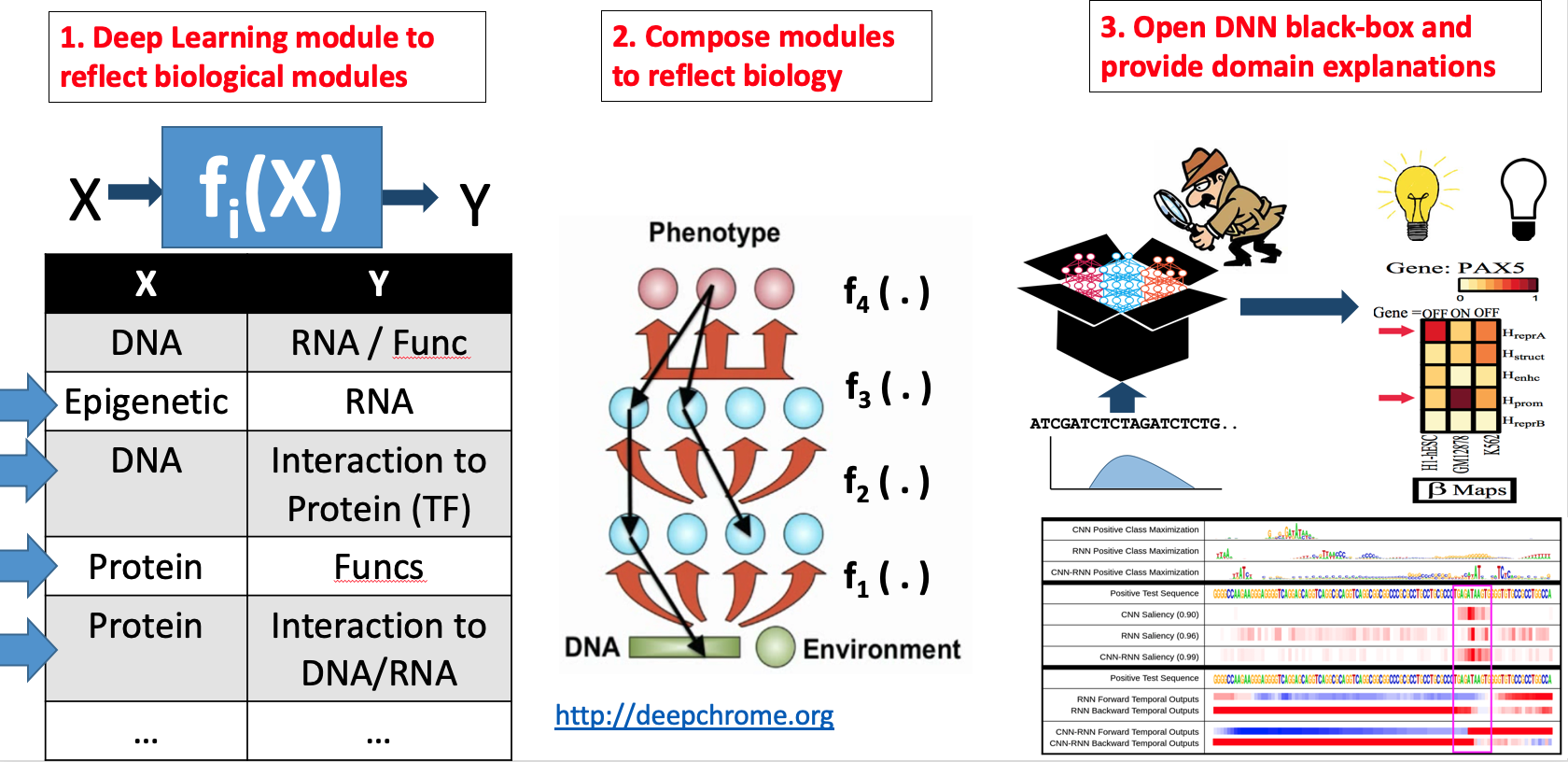
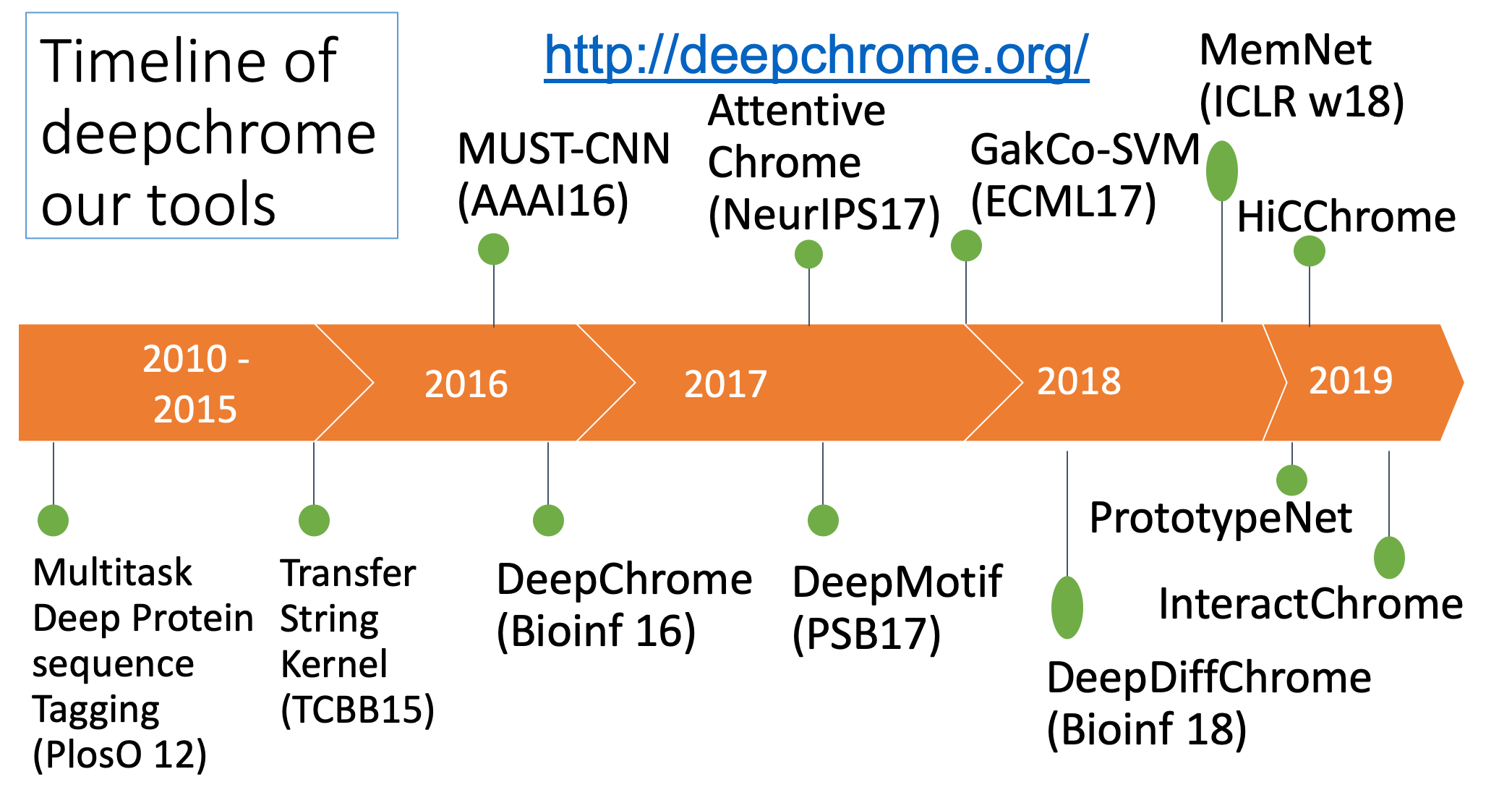
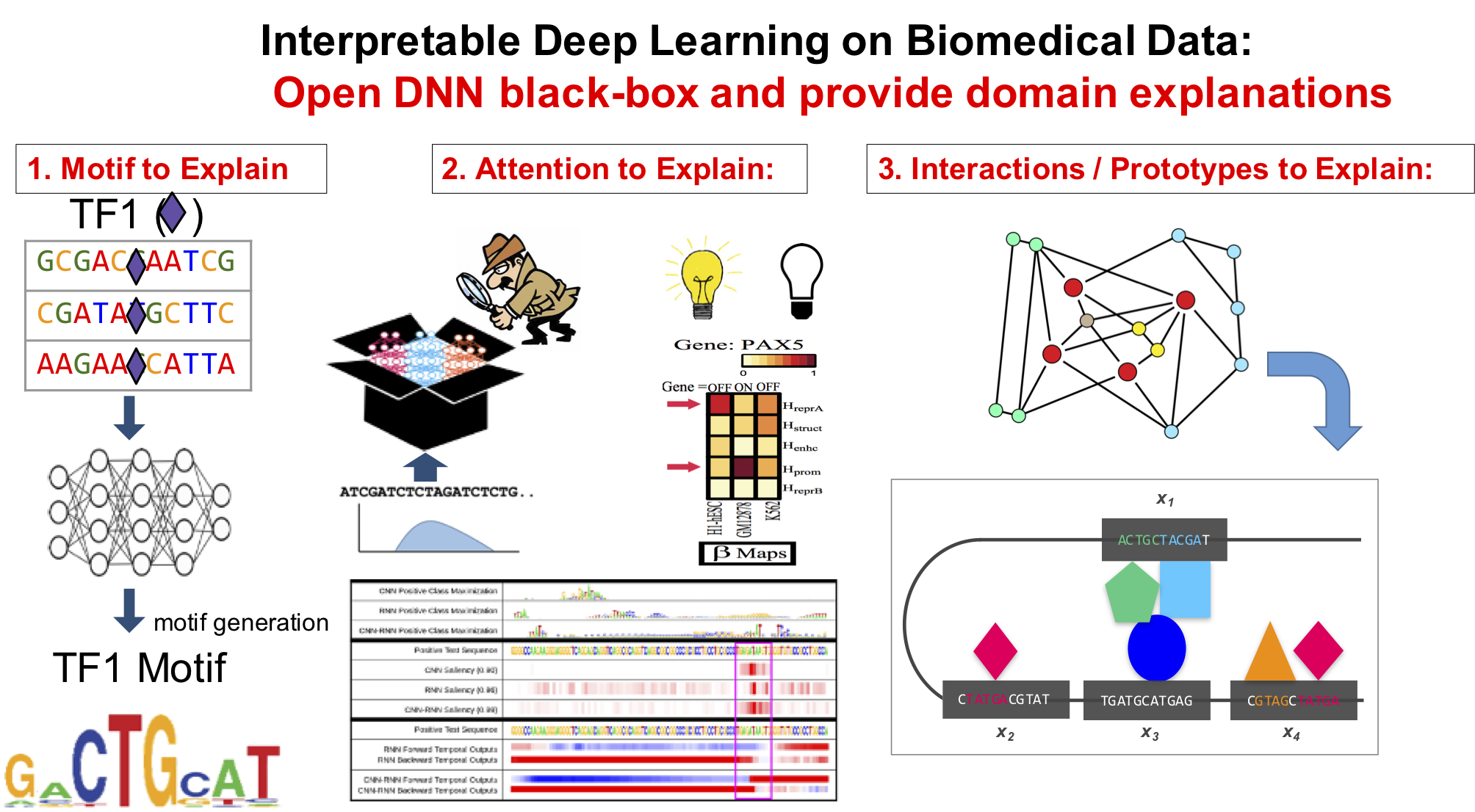
- "Attend and Predict: Understanding Gene Regulation by Selective Attention on Chromatin", at (NeurIPS 2017 ) (PDF)
-
"DeepDiff: Deep-learning for predicting Differential gene expression from histone modifications", Bioinformatics 2018 (PDF)
-
"DeepChrome: Deep-learning for predicting gene expression from histone modifications", Bioinformatics 2016 (PDF)
- "Memory Matching Networks for Genomic Sequence Classification", ICLR2017, (Arxiv)
(Poster)
- "GaKCo: a Fast GApped k-mer string Kernel using COunting", ECML17, (Arxiv)
-
"Deep Motif Dashboard: Visualizing and Understanding Genomic Sequences Using Deep Neural Networks", PSB17 (Arxiv)
-
"Deep Motif: Visualizing Genomic Sequence Classifications", ICLR2016, the International Conference on Learning Representations (Arxiv)
- "MUST-CNN: A Multilayer Shift-and-Stitch Deep Convolutional Architecture for Sequence-based Protein Structure Prediction", AAAI 2016. (PDF) (Talk)
- "Unsupervised Feature Learning by Deep Sparse Coding", SDM 2014. (PDF)(Talk)
- "Deep Learning for Character-based Information Extraction",ECIR 2014, (PDF)
(SupplementaryDoc)(Talk)
- "Learning the Dependency Structure of Latent Factors",NIPS 2012, (PDF)(Talk)
- "A unified multitask architecture for predicting local protein properties", PLoS ONE 2012((PDF))
- "Sentiment Classification with Supervised Sequence Encoder", ECML 2012, (PDF)(Talk)
- "Sentiment Classification Based on
Supervised Latent n-gram Analysis",CIKM 2011, (PDF)(Talk)
- "Semi-Supervised
Abstraction-Augmented String Kernel for Multi-Level Bio-Relation
Extraction", ECML 2010, (PDF)(Talk)
- Semi-Supervised Bio-Named Entity Recognition with Word-Codebook
Learning", SDM 2010, (PDF)
- "Polynomial Semantic Indexing", NIPS 2009, (PDF)
- "Semi-Supervised Sequence Labeling with Self-Learned Feature", ICDM 2009, (PDF)(Talk)
- "Combining Labeled and Unlabeled Data for Word-Class Distribution" , CIKM 2009, (PDF)
|
Learning Graph from Data
(Helping researchers effectively translate aggregated data into knowledge that take the form of graphs)
[ Our tools for learning networks from heterogeneous samples (by joint structure learning of Gaussian Graphical Models)]
|
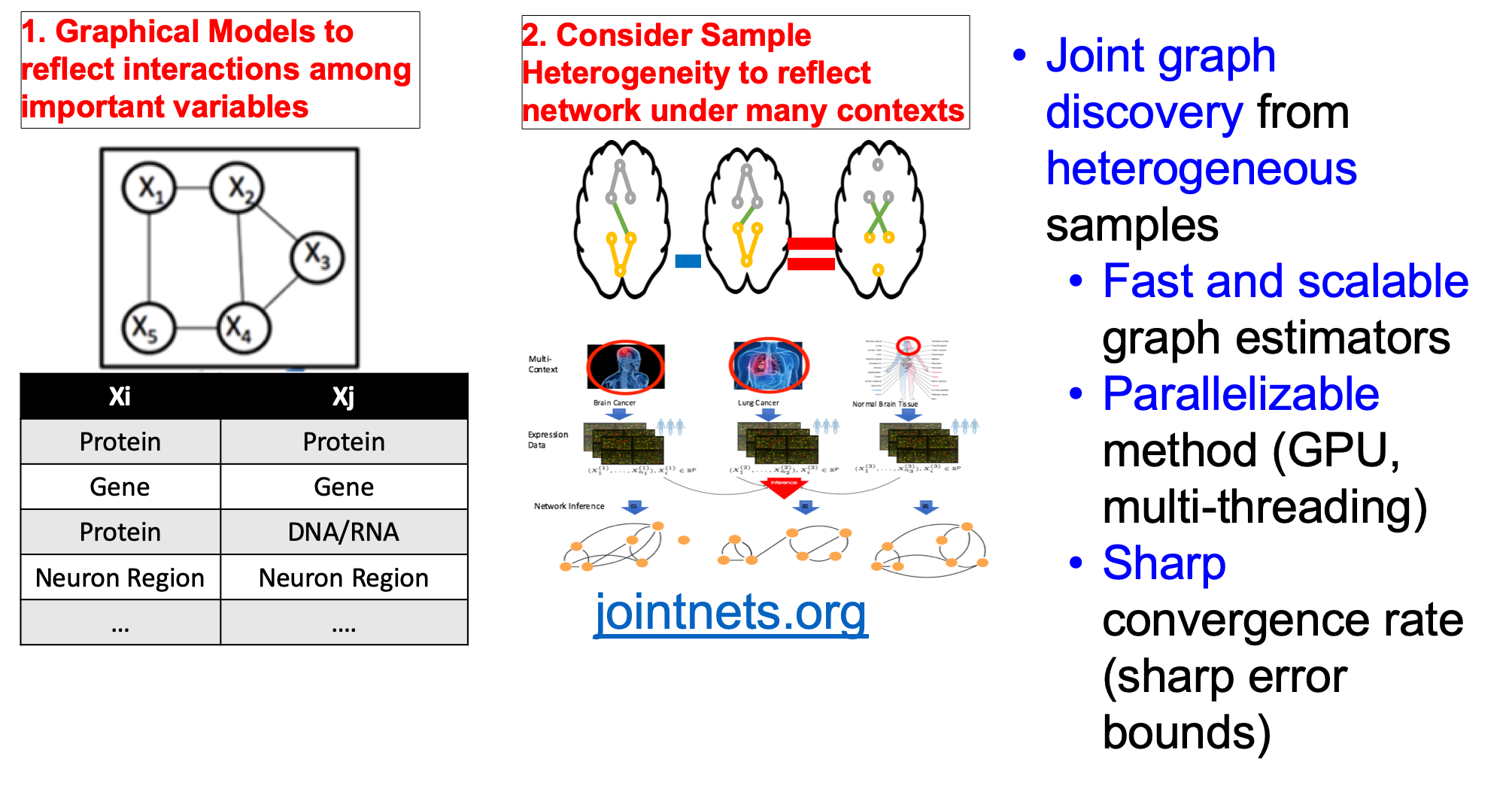
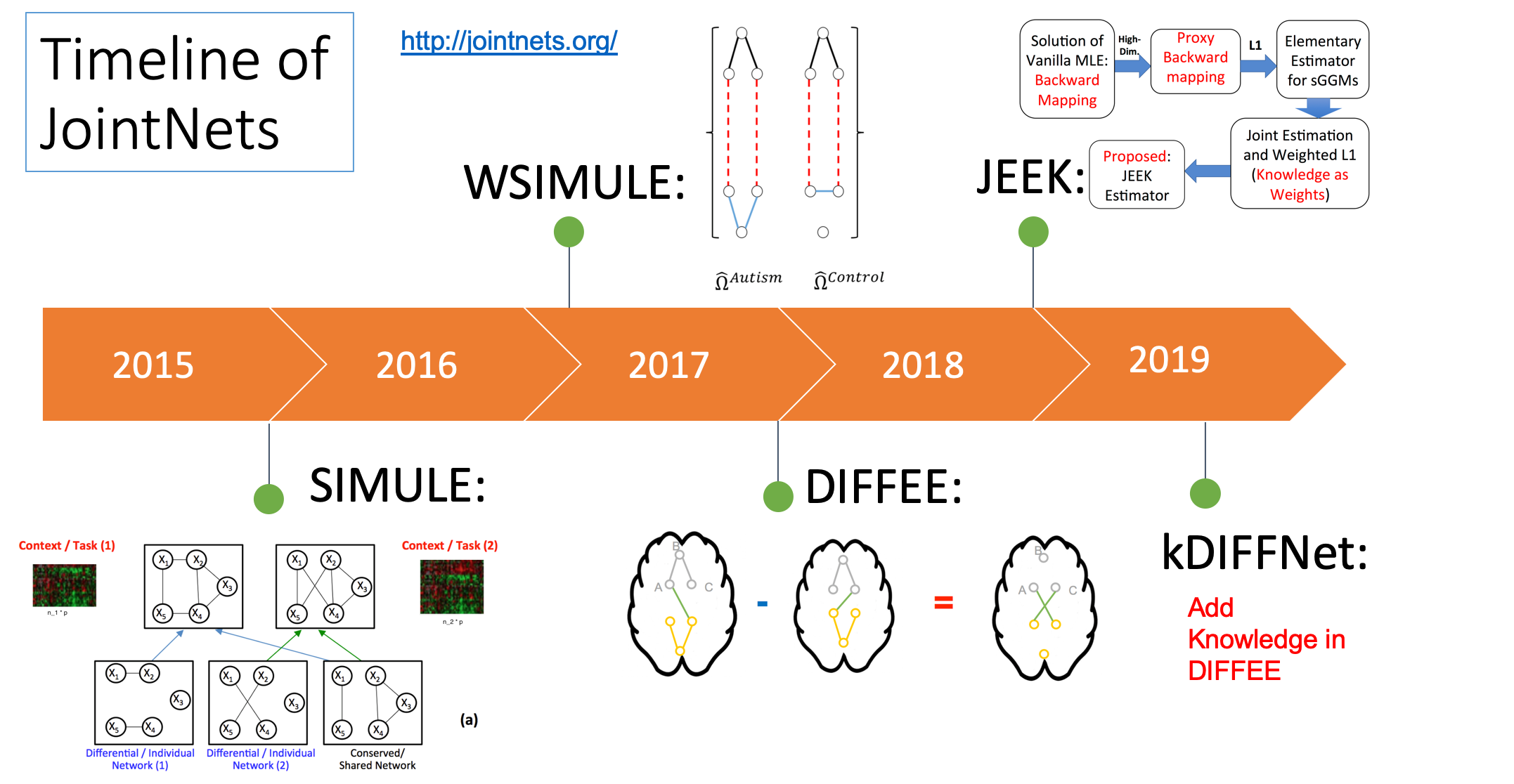
- "A fast and scalable joint estimator for integrating additional knowledge in learning multiple related sparse gaussian graphical models.", ICML (2018)(PDF)
- "Fast and Scalable Learning of Sparse Changes in High-Dimensional Gaussian Graphical Model Structure", AISTATS (2018)(PDF)
- "A Fast and Scalable Joint Estimator for Learning Multiple Related Sparse Gaussian Graphical Models", AISTATS (2017)(Arxiv)(PDF)
- "A constrained L1 minimization approach for estimating multiple Sparse Gaussian or Nonparanormal Graphical Models", (2017) Machine Learning Journal ; (PDF
)
- "Learning the Dependency Structure of Latent Factors",NIPS 2012, (PDF)(Talk)
- "Transfer String Kernel for Cross-Context DNA-Protein Binding Prediction",@ (TCBB) (Arxiv)
- "Protein Complex Identification by Supervised Graph Clustering", Bioinformatics 2008, (PDF)(Talk)
|
Add Weak Supervision in Machine Learning |
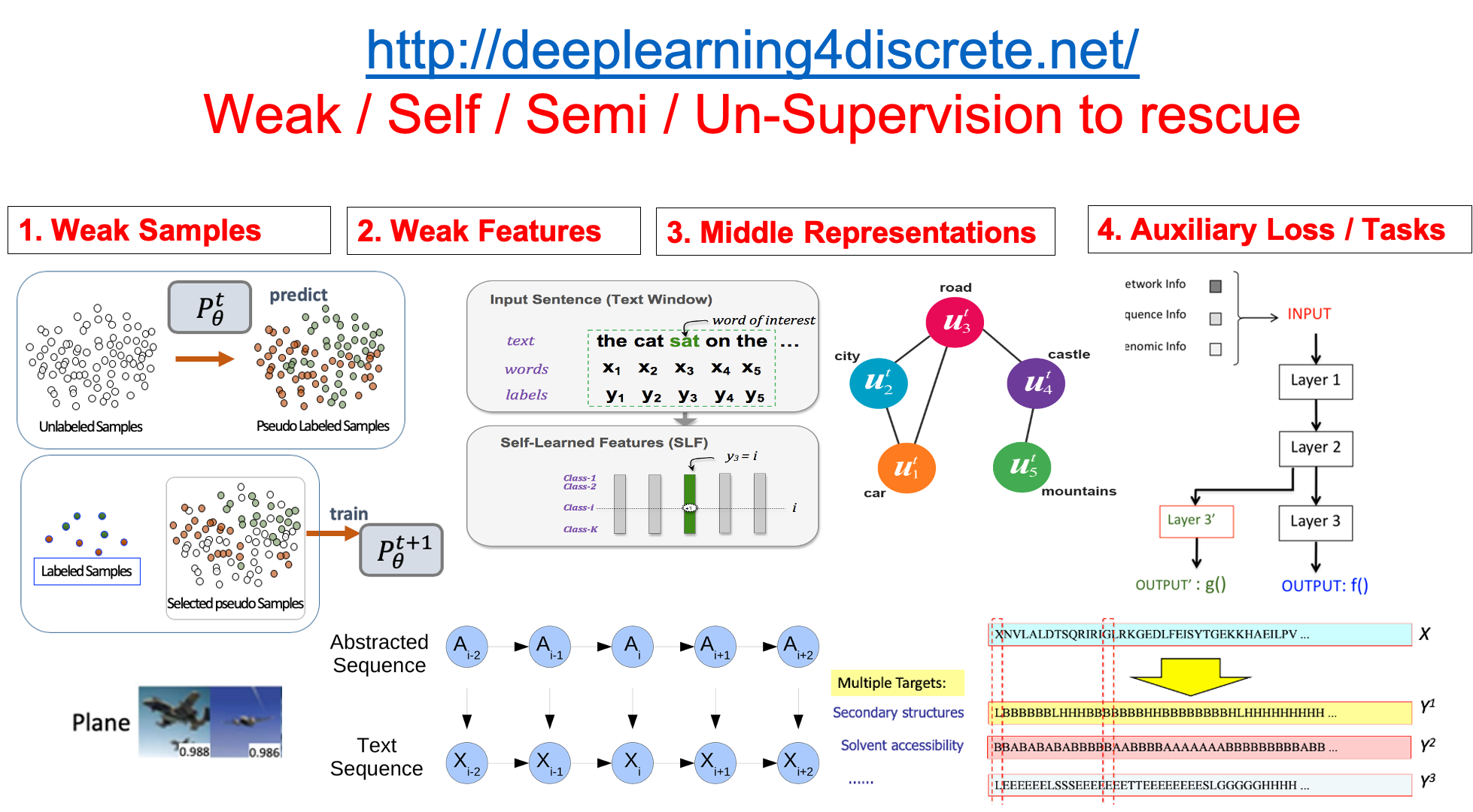
- "Curriculum Labeling: Self-paced Pseudo-Labeling for Semi-Supervised Learning", P Cascante-Bonilla, F Tan, Y Qi, V Ordonez, arXiv:2001.06001
- " Yanjun Qi, B. Bai, X. Ning, and P. Kuksa. Systems and methods for Semi-supervised Relationship Extraction, Apr. 2014.
- "Semi-Supervised Convolution
Graph Kernels for Relation Extraction", SDM 2011,
(PDF) (Talk)
- "Semi-Supervised Multi-Task Learning for Predicting Interactions between HIV-1 and Human Proteins", ECCB 2010, (PDF)
(Talk)
|
Machine Learning for Health |
- "Kernelized Information-Theoretic Metric Learning for Cancer Diagnosis using
High-Dimensional Molecular Profiling Data", TKDD 2016, (PDF)
- "MAPer: A Multi-scale Adaptive Personalized Model for Temporal Human Behavior Prediction ",CIKM 2015,
(PDF)
(Talk)
- "Causal Analysis of Inertial Body Sensors for Enhancing Gait Assessment Separability towards Multiple Sclerosis Diagnosis", IEEE Body Sensor Network (BSN) 2015, (PDF) (Talk)
- "Piecewise Linear Dynamical Model for Action Clustering from Real-World Deployments of Inertial Body Sensors", BodyNets 2014, (PDF) (Talk)
- Retrieving Medical Records with sennamed: NEC Labs America at TREC 2012 Medical Record Track, 2012 Text Retrieval Conference, (PDF)
|








